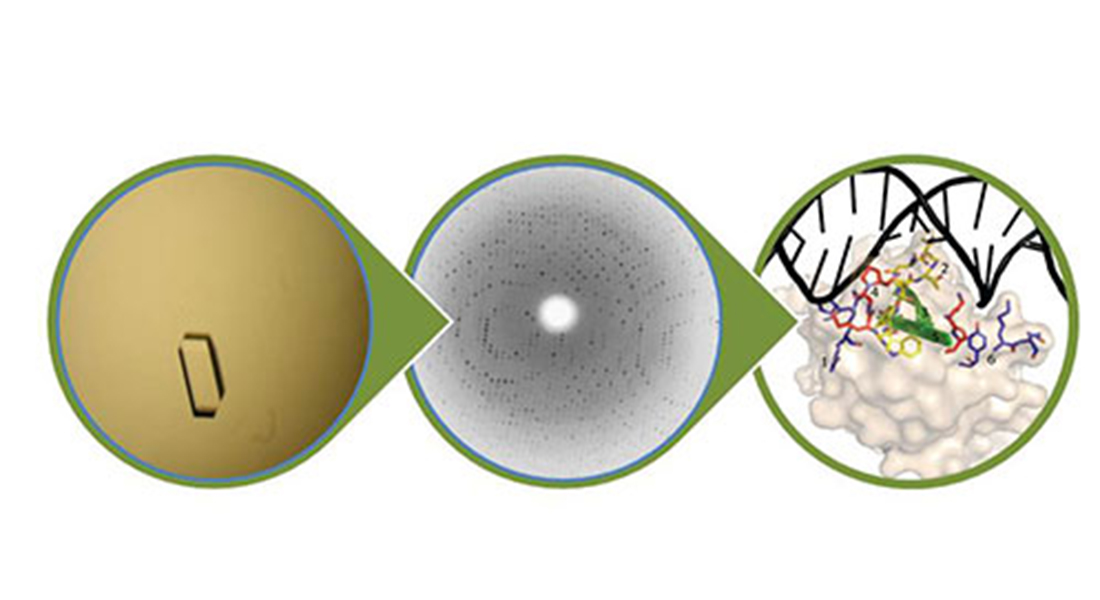
Crystallography - Lo Leggio Group
Crystallography is the primary technique used in atomic structure determination of molecules of all sizes, from simple chemical compounds to large biological macromolecular complexes.
My group works on macromolecules structure and function, primarily proteins, in particular enzymes, and their interactions with each other and other macromolecules (nuclei acids and polysaccharides).
Enzymes involved in polysaccharide biomass degradation and modification
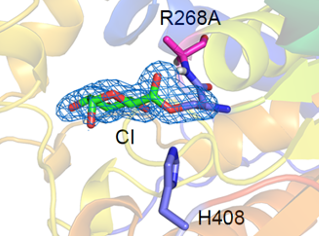
Biomass is an important renewable resource, and much of it is composed by polysaccharides, especially when coming from plants, like in food or agricultural waste. These polysaccharides are often crystalline and embedded in complex matrices also involving other components like lignin, leading to high recalcitrance to breakdown. This is an obstacle for using renewable resources for producing eg bioethanol or biomaterials. In my group we study the mechanisms of enzymes involved in plant biomass degradation at the atomic level, as well as their interactions with complex plant biomass components. We study for example glycoside hydrolases, lytic polysaccharide monooxygenases and glucuronoyl esterases. Below an example of a covalent intermediate trapped in a variant of a glucuronyl esterase, and enzyme involved in decoupling lignin and polysaccharides. In the paper, part of an ongoing collaboration with Johan Larsbrink at Chalmers University of Technology, we use crystallography, biochemistry and computation to better understand the mechanism and learn to utilize these enzymes beneficially.
Lysogeny-lysis switches in temperate bacteriophages
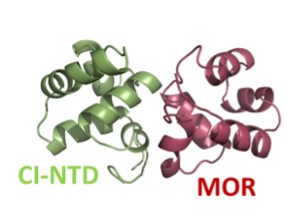
The other main research theme in my lab are DNA binding transcription factors, particularly the ones involved in lysogeny-lysis switches. These switches decide whether temperate phages go into the lysogenic or lytic cycle (killing the bacteria) and they are important in transfer of genetic material between bacteria. We are particularly interested in switches involving the CI-MOR protein-protein interaction shown below, particularly the ones from Gram-positive pathogenic bacteria bacteriophages (like the Staphylococcus aureus phi13 phage). In these projects we collaborate with other structural biologists and microbiologists.
Advanced macromolecular crystallography
Although we use many techniques, our favourite is crystallography. Aside from ‘just’ determining structures of proteins and their interaction with small and large molecules as in the example above, we also use more advanced crystallography. For example, high resolution crystallography to determine the protonation state of residues in enzymes (read more here) or our recent detailed work on X-ray induced photoreduction of lytic polysaccharide monooxygenases (read more here). We are avid users of synchrotron facilities like MAX IV and the ESRF and cooperate with several facility scientists. I also work very closely with the other crystallographers in the Biological Chemistry Section (Anders Kadziola, Pernille Harris, Sine Larsen)
I am also a member of ISBUC and the Centre for Medicinal Chemistry at the University of Copenhagen.
Student projects are available in all areas of research mentioned under 'RESEARCH' and several other collaborative projects with other groups. Additionally to their own research projects, the crystallography group supports the synthetic chemists in the Department in characterization of the compounds.
I am currently funded by the Novo Nordisk Foundation and the Independent Research Fund Denmark.
Associated Researchers
| Name | Title | Phone | |
|---|---|---|---|
| Anders Kadziola | Associate Professor | ||
| Leila Lo Leggio | Professor | +4535320295 |
