Protease Specificity and Function
CECB has developed a range of technologies to monitor and determine protease specificity and inhibition. Protease research serves to investigate the physiological significance of the proteases and to determine the when, the where and the why of proteolytic activity. A unique combinatorial chemistry toolbox for the investigation of enzymes has been developed at the Center for Evolutionary Chemical Biology and is employed during investigation of the mamalian proteases. This toolbox is employed to investigate the specificity of proteases in a variety of live cells and tissue. Genes encoding for proteases involved in plant germination, arthritis and in programmed cell death are cloned and expressed.
CECB has established a platform with bioinformatics, modelling, molecular biology, protein expression, FRET, enzymology, combinatorial chemistry, synthesis, high-throughput screening, in vivo substrates and microscopy. These tools are used to identify the genes, express the proteases, analyze the protease activity profile, locate and study the proteases in vivo in a time- and tissue-specific manner and identify the natural target for the protease.
Caspase 8: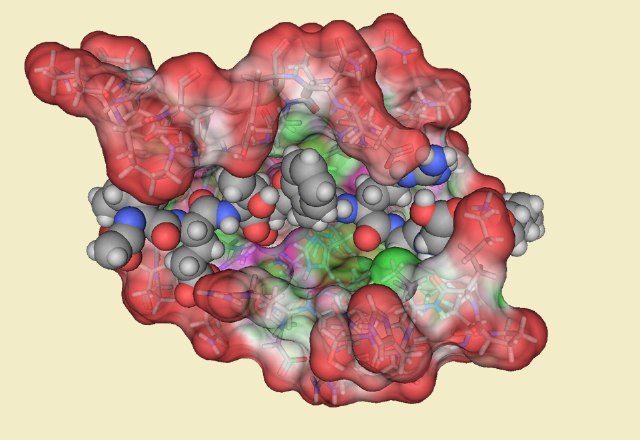 Carboxypeptidase B:
Carboxypeptidase B: 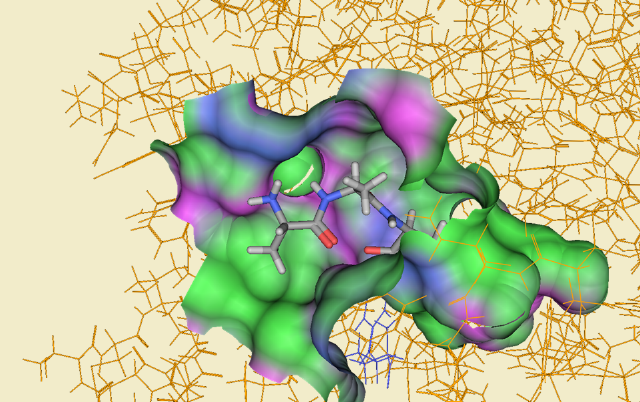
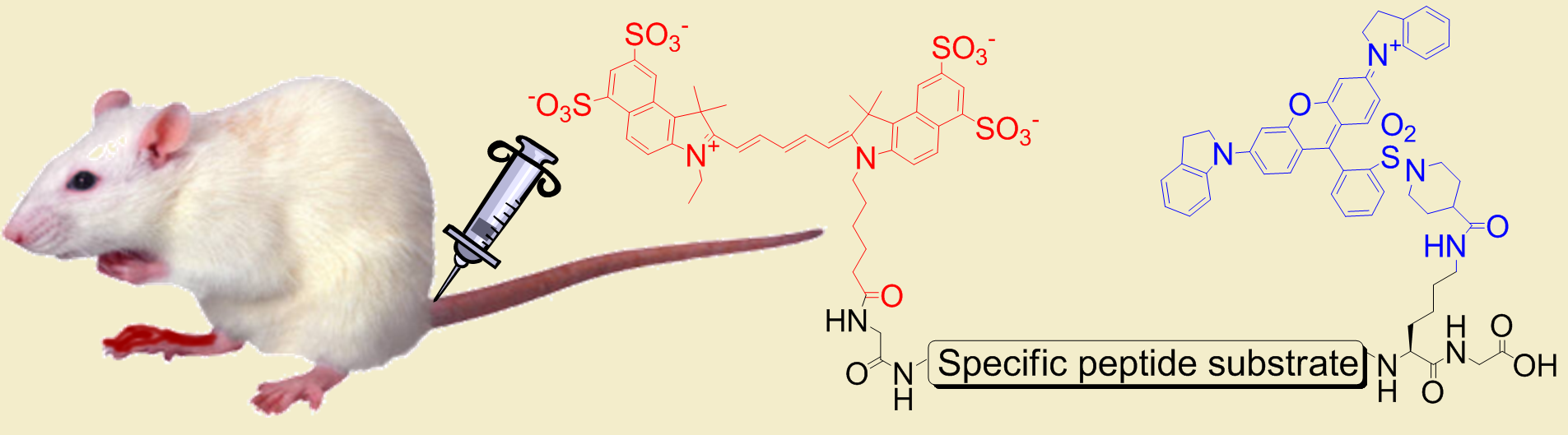
Artificially induced arthritis may be monitored using specific in vivo probes that can be visualized through intact tissue. Fluorescence increases selectively at the site of inflammation.
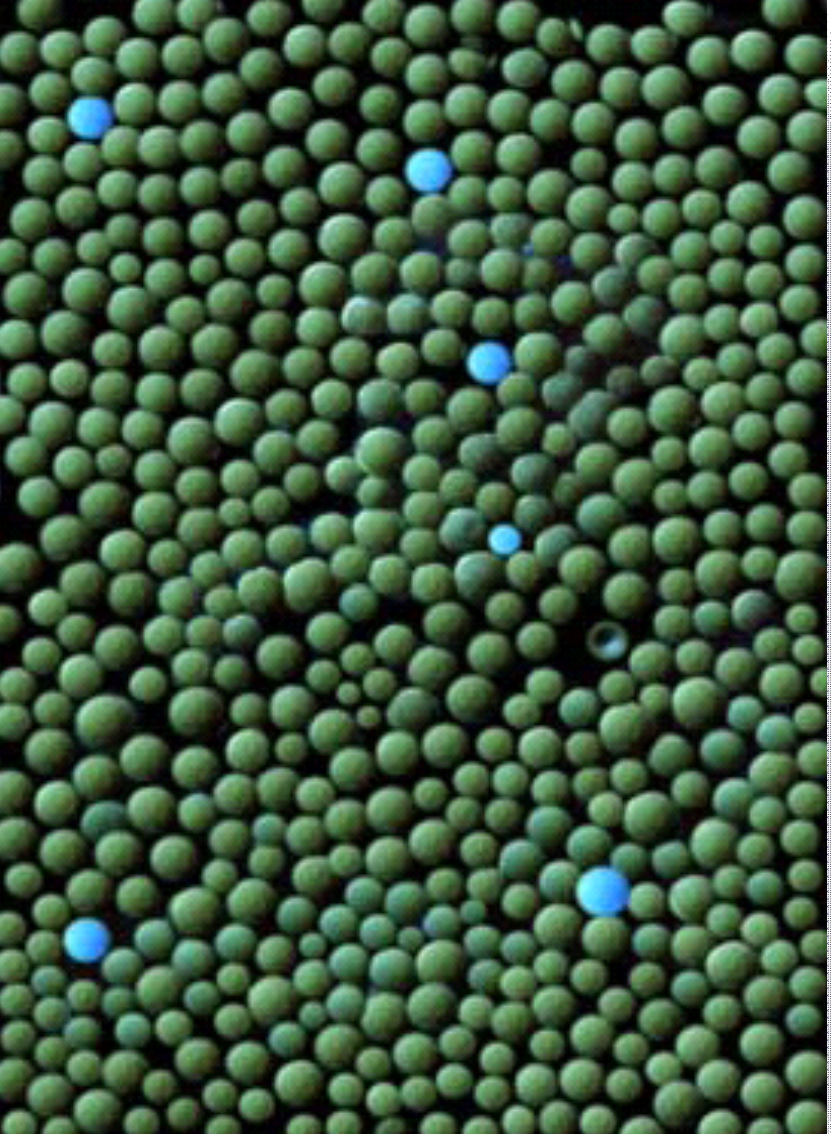
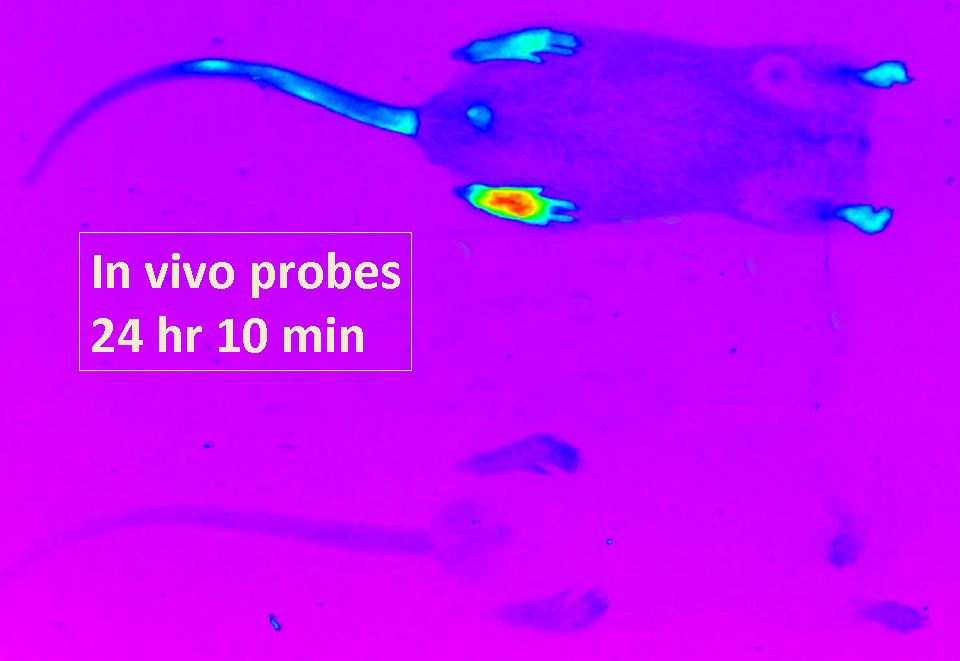
Substrate specificity of a target protease over that of related proteases is determined through screening of combinatorial libraries. The most selective substrates are converted into in vivo probes for live monitoring of disease progression.
